Predicting Student Success
Why do some students graduate and some students not graduate? What are the best predictors of graduation? Which courses are the most essential to succeeding in a given major, and how early should they be taken? How and why do students choose their majors? What makes a student decide to change majors? Are there early-on interventions that can be made to help students succeed? What are the roadblocks to student success?
This research uses data mining and machine learning techniques.
Computational Morphodynamics
Computational Morphodynamics may be defined as the study of the three-way interaction of physical, informational, and geometrical processes that influences the changing form, shape, and structure of living cells, tissues and organisms.
Some of the scientific questions we ask include:
- How is the morphology of organisms specified?
- How do mechanics, cell growth, and cell division affect morphology?
- How do biochemical and informational processes determine the major changes in the morphology of living organisms?
- What sort of physics is necessary to describe morphological development, and how does it feed back into the traditional biological and biochemical processes?
- Can we design and build synthetic biological systems to mimic various aspects of morphological development?
- What are the computational consequences of the macroscopic patterns of mammalian brain cell development?
- How do living morphodynamic systems evolve?
- How can we design self-fabricating molecular structures?
- What kinds of computational processes are best described using morphodynamics?
- How can we classify morphodynamic systems, up to Turing-computable diffeomorphisms?
- What sort of computational and mathematical resources are necessary to really understand morphology, morphological development, and morphological dynamics?
As scientists we seek to understand how morphology is specified and changes over time.
The term computational morphodynamics itself pays tribute to the diversity of disciplines the subject depends on: morphology, dynamics, and computation.
But more than that, it reminds us that the study of morphological development lies at the interface between biology, mathematics, computation, and engineering.
These disciplines give us the tools we need to actually do good science. Things like mechanics, growth, cell division, and biochemical and bio-mechanical regulation and feedback mechanisms certainly contribute to morphological dynamics. Understanding how each process works and how they relate to one another is far from trivial. We believe that by utilizing and integrating these multidisciplinary tools we can obtain more comprehensive and specific answers to the questions we pose.
Caltech Biological Network Modeling Center (BNMC)
 The goal of the BNMC is to bring together
Caltech biologists, bioengineers, mathematicians, and
computer scientists to develop and apply state-of-the-art
computational tools for modeling and analyzing complex
biological systems. The focus of the BNMC is biochemical
phenomena occurring within and between cells, in particular
the mechanistic modeling of molecular networks of all kinds
(e.g., transcriptional, regulatory, metabolic, signal
transduction, mechanical, etc.) with and without spatial
variation and intercellular communication. This is at once
one of the oldest areas of modeling in biology and one that
continues to challenge theoreticians and biologists alike.
The BNMC is funded by Caltech's Beckman Institute.
The goal of the BNMC is to bring together
Caltech biologists, bioengineers, mathematicians, and
computer scientists to develop and apply state-of-the-art
computational tools for modeling and analyzing complex
biological systems. The focus of the BNMC is biochemical
phenomena occurring within and between cells, in particular
the mechanistic modeling of molecular networks of all kinds
(e.g., transcriptional, regulatory, metabolic, signal
transduction, mechanical, etc.) with and without spatial
variation and intercellular communication. This is at once
one of the oldest areas of modeling in biology and one that
continues to challenge theoreticians and biologists alike.
The BNMC is funded by Caltech's Beckman Institute.
Cell Simulation and The Computable Plant
The ultimate fate of all living cells is
regulated by a complex set of interactions between various
proteins and nucleic acids. Some proteins act directly as
transcription factors that control the expression of one or
more genes. Others interact with one another along a
diverse sequence of pathways that eventually act to either
increase or decrease the transcription of specific genes.
We are currently developing a new mathematical and
computational infrastructure to characterize these pathways
and the interactions between these complex molecules and
the cellular environment. The goal is to develop a model
system based on known or inferred biological data that
addresses such questions as how the various regulators are
regulated, to what extent these molecules are affected by
the cellular environment, and what the effect of the
inactivity or over-activity of one or more of these
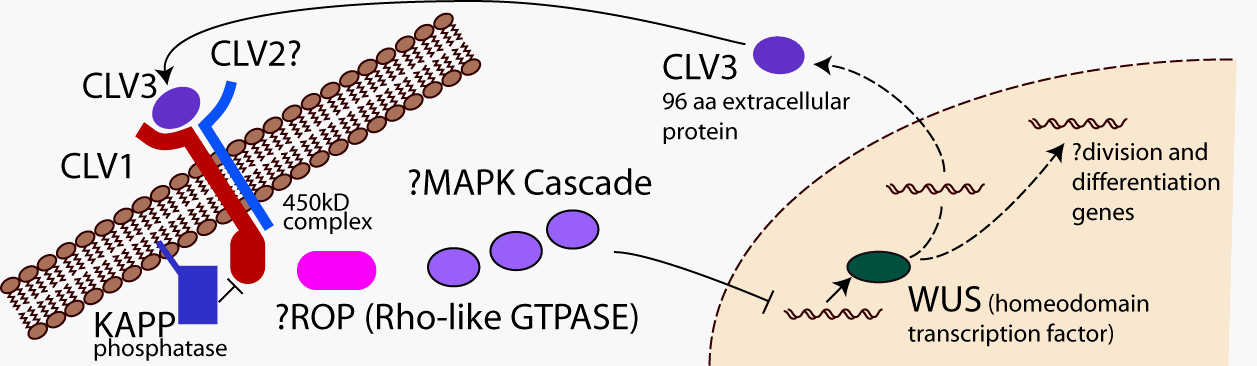
molecules is or should be. 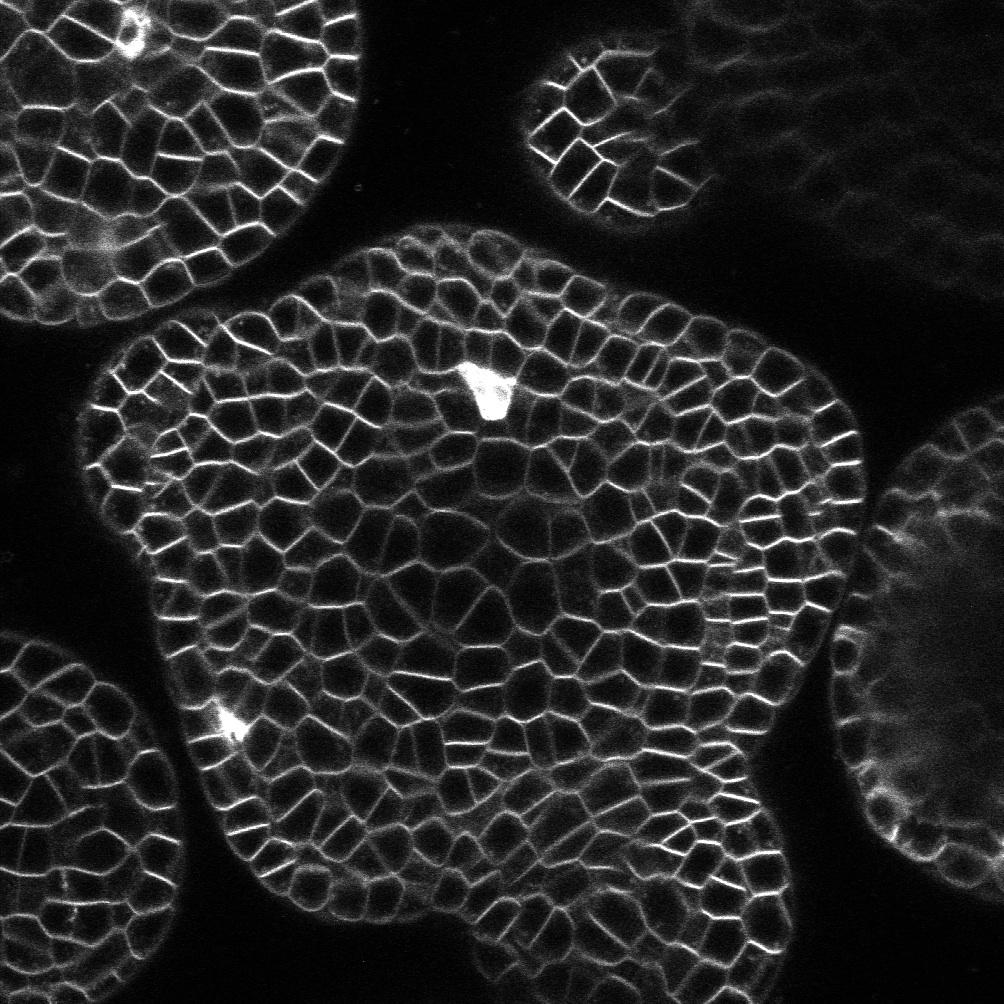
Issues that must be considered while developing this infrastructure include the mathematical formulation of large-scale molecular interactions; computational methods to describe the molecular interactions and developmental pathways; how to acquire, maintain, and archive the necessary biological data in a computational environment, including an interactive data management system with hypertext links to published results; and investigator user-interfaces for simulation and prediction.
 The computable plant project focuses
on a quantitative description of plant development. Key
elements include a mathematical (ODE-based) framework
combining transcriptional regulation, signal transduction,
and dynamics; computer algebra based model generation
(Cellerator); automated code generation; nonlinear
optimization; automated image processing and data mining;
and numerical simulation. Fitting predictive models to
high-resolution Arabidopsis thaliana image data allows
in-silico exploration (via simulation) of alternative
scientific hypotheses and guides in experiment design.
Collaboration with Mjolsness Lab at UC, Irvine; Meyerowitz
Lab at CalTech; and an international team of
researchers.
The computable plant project focuses
on a quantitative description of plant development. Key
elements include a mathematical (ODE-based) framework
combining transcriptional regulation, signal transduction,
and dynamics; computer algebra based model generation
(Cellerator); automated code generation; nonlinear
optimization; automated image processing and data mining;
and numerical simulation. Fitting predictive models to
high-resolution Arabidopsis thaliana image data allows
in-silico exploration (via simulation) of alternative
scientific hypotheses and guides in experiment design.
Collaboration with Mjolsness Lab at UC, Irvine; Meyerowitz
Lab at CalTech; and an international team of
researchers.
Click here to vist the Computable Plant Web Site
Cellerator and xCellerator
 Cellerator is a mathematica package that
describes single and multi-cellular signal transduction
networks (STN) with a compact optionally palette-driven,
arrow based notation to represent biochemical reactions and
transcriptinoal activation. Multi- compartment systems are
represented as graphs with STNs embedded in each node.
Interactions include mass- action, enzymatic, allosteric
and connectionist models. Reactions are symbolically
translated into differential equations and can be solved
numerically to generate predictive time courses or output
as systems of equations that can be read by other programs.
Cellerator is freely downloadable for
academic/government/nonprofit users.Click here to visit the
Cellerator web page.
Cellerator is a mathematica package that
describes single and multi-cellular signal transduction
networks (STN) with a compact optionally palette-driven,
arrow based notation to represent biochemical reactions and
transcriptinoal activation. Multi- compartment systems are
represented as graphs with STNs embedded in each node.
Interactions include mass- action, enzymatic, allosteric
and connectionist models. Reactions are symbolically
translated into differential equations and can be solved
numerically to generate predictive time courses or output
as systems of equations that can be read by other programs.
Cellerator is freely downloadable for
academic/government/nonprofit users.Click here to visit the
Cellerator web page.
xCellerator is similar to Cellerator but the implementation is fundamentaly different; it consequently runs 100 - 1000 times faster then Cellerator. It is currently in alpha-test stage, and does not havel all features implemented. When completed, it will be fully backwards compatible with Cellerator models, but will be more flexible and compatible with other programs. It will also be fully integrated with MathSBML. Visit the xCellerator web page.
Both Cellerator and xCellerator require Mathematica.
Systems Biology Markup Language (SBML)
 The
Systems Biology Markup Language (SBML) is a model
representation formalism for system biology, allowing
computer programs to communicate with one another using a
standardized file format. SBML is oriented towards
describing systems of biochemical reactions such as those
that arise in the study of signal transduction networks. It
is based on XML (the
eXtensible Markup Language). Click here to visit the
SBML web page.
The
Systems Biology Markup Language (SBML) is a model
representation formalism for system biology, allowing
computer programs to communicate with one another using a
standardized file format. SBML is oriented towards
describing systems of biochemical reactions such as those
that arise in the study of signal transduction networks. It
is based on XML (the
eXtensible Markup Language). Click here to visit the
SBML web page.
MathSBML
The MathSBML package for Mathematica provides facilities for reading models expressed in SBML, converting them to systems of ordinary differential equations, and solving and plotting the results in Mathematica. MathSBML is open source software (LGPL license) and can be freely downloaded form SourceForge.
Microgravity Transcriptome*
 Alterations in
gravity impact cells in many ways, including cell
proliferation, chromosomal aberations, gene expression, and
processes of reproductive cell formation. At the systemic
level, microgravity affects virtually every system in the
body. Known detremental changes include bone mineral
density losses that vary across specific bone sites with
greater loss in the weight-bearing bones. Marked atrophy of
postural muscles as well as reduced muscle strength and
tasks prevent astronauts from performing sustained work in
space and cause severe side-effects upon return to Earth.
These effects increase with time in flight and it is not
known how long they continue to develop. e are studying the
changes in gene expression due microgravity on muscle and
bone tissue due to microgravity in an attempt to determine
the signaling pathways involved in these changes. This
project will help to understand the molecular and cellular
causes of muscle atrophy and identify potential targets for
counteremasure. Collaboration with the Johnson Space Center
and the Wold Lab at CalTech.
Alterations in
gravity impact cells in many ways, including cell
proliferation, chromosomal aberations, gene expression, and
processes of reproductive cell formation. At the systemic
level, microgravity affects virtually every system in the
body. Known detremental changes include bone mineral
density losses that vary across specific bone sites with
greater loss in the weight-bearing bones. Marked atrophy of
postural muscles as well as reduced muscle strength and
tasks prevent astronauts from performing sustained work in
space and cause severe side-effects upon return to Earth.
These effects increase with time in flight and it is not
known how long they continue to develop. e are studying the
changes in gene expression due microgravity on muscle and
bone tissue due to microgravity in an attempt to determine
the signaling pathways involved in these changes. This
project will help to understand the molecular and cellular
causes of muscle atrophy and identify potential targets for
counteremasure. Collaboration with the Johnson Space Center
and the Wold Lab at CalTech.
Treatment-related Leukemia*
Traditional therapies for lymphoma -- such as high dose chemo/radiotherapy following by autologous peripheral blood stem cell transplantation -- are often associated with the development of secondary cancers -- known as treatment-related myelodysplasia or acute myeloid leukemias (t-MSDS/AML). Operating under the hypothesis that pre-transplant and transplant-related chemo/radiotherapeutic exposures induce detectable hematopoietic and genetic abnormalities that antedate and predict for clonal evolution and development of t-MDS/AML, we are using RNA micro-array, data-mining, and bioinformatic modeling in an attempt to determine the genetic regulatory network and relevant proteins involved. Collaboration with the City of Hope National Medical Center. Sponsored by NASA and NCI.
*This research was funded approximately 2001-2005, primarily by NASA. The organizations that provided support no longer exist and this is no longer an area of active research for me.
Spreading Depression
Associated with migraine headache, stroke, concussion, seizure, and transient global amnesia, spreading depression (SD) is a slowly moving reduction (hence the term "depression") of electrical (EEG) activity in the brain. The SD wave, which travels at 3 - 12 mm/minute and can last for up to 2 minutes at any point, consists of membrane depolarization and ionic concentration changes, is usually preceded by epileptic-like electrical burstis and is typically followed by increased bllod flow and a prolonged period of vasodilation. I have developed a computational physiological model that describes some of the basic mechanisms involved in SD.
Theory of Force Probe Microscopy
Force measurements on and within single macromolecular complexes utilizing such techniques as atomic force microscopy (AFM), optical trapping, flexible glass fibers, and magnetic beads provide a rich source of quantitative data on biomolecular processes. These experiments typically measure the macromolecular binding forces between single protein-ligand pairs
Stochastic thermal fluctuations, an undesirable source of noise in macroscopic biochemical experiments, are an essential element of these sensitive and novel experiments. With the proper analysis, a great deal of information can be gleaned from measurements of these fluctuations. A quantitative framework for analyzing such measurements, based on Kramers’ theory of molecular dissociation, has been developed developed. The analysis reveals the kinetic origin and stochastic nature of the measurements.
This analysis was performed during the late 1990's and I am no longer actively engaged in research in this area.
Satellite Navigation*
Exact Repeat Ground Track Orbit Design and Maintenance. The ground track of a satellite orbit is the locus of points traced out on the Earth's surface directly beneath the spacecraft. Earth observation satellites (such as TOPEX/Poseidon, Jason, LANDSAT, GEOSAT, Quickscat and SEASAT) are typically placed in orbits with ground tracks that repeat after a fixed number of days. These low-earth orbiting satellites typically have a period of around two hours. During this time the Earth rotates by approximately 30 degrees (360˚ in 24 hours). Since the plane of the satellite orbit rotates much more slowly (typically one or two degrees a day), if the ground track crosses the equator at some longitude L on one orbit, it will cross the equator around (but not precisely) 30˚ further west on the next orbit. If the total westward change after D days and N orbits is an integral muliple of the difference between the Earth's angular rate (W=360˚/24 hours) and the satellite's orbital plane rotation rate (R), the ground track will exactly repeat. To place (and maintain) a satellite in such an orbit, it must therefore have a period of (360 D/N) (W-R) (Note: in standard notation, W is little omega sub e, and R is the derivative with respect to time of capital omega, the right ascension of ascending node).
Frozen Orbits. Earth observation missions are frequently placed in low eccentricity forzen orbits with the perigee fixed at 90˚. The such an orbit, the mean argument of perigee, w (lower-case greek letter omega), and the the mean eccentricity, e, are kept in the neighborhood of a stable critical point. Deviations from the critical point lead to closed curves in the (e, w) phase plane when only central-body gravitational perturbations are considered. These trajectories remain in the neighborhood of the critical point even under the influence of drag and solar radiation pressure, but the phase trajectories are far more complicated. Existance of the frozen orbit is usually attributed to a balancing of the secular perturbations of even zonal harmonics with the long period perturbations of the odd zonal harmonics.
GTARG*
GTARG is the Ground Track Maintenance Maneuver Targeting Program developed for exact repeat ground track orbiting satellites. The GTARG algorithm propagates non-singular mean elements using Merson's extension of Grove's theory for the geopotential field through J30; Kaula's disturbing function for luni-solar gravity; and a Jacchia-Roberts atmosphere.
*This work was supported by NASA through the late 1990's and I am no longer working on this area.

